Marine resource use has influenced human population on the Central American Isthmus for millennia
Micropaleontology
Modern computational tools
may open a new era for
fossil pollen research
Western Africa and Northern South America
By integrating machine-learning technology with high-resolution imaging, scientists are improving the taxonomic resolution of fossil pollen identifications and greatly enhancing the use of pollen data in ecological and evolutionary research.
One of the best sources of information on the evolution of terrestrial ecosystems and plant diversity over millions of years is fossil pollen. For palynologists—the scientists who study ancient pollen—a common challenge in the field is the identification of plant species based on these fossil grains. By integrating machine-learning technology with high-resolution imaging, a team from the Smithsonian Tropical Research Institute (STRI), the University of Illinois at Urbana-Champaign, the University of California, Irvine and collaborating institutions was able to advance this goal.
Pollen is resistant to disintegration, even when exposed to elevated temperatures or strong acids. This allows it to preserve in sediments for millions of years, making it an invaluable record of how the different groups of plants have evolved and the environmental factors that have played a role in this evolution. In order to achieve this, researchers must first be able to identify what they are observing under the microscope.
Morphological variation of the pollen type named as Striatopollis catatumbus. Credit: Ingrid Romero and Michael Urban.
To help improve the efficiency and accuracy of fossil pollen identifications, scientists developed and trained three machine-learning models to differentiate among several existing Amherstieae legume genera and tested them against fossil specimens from western Africa and northern South America dating back to the Paleocene (66–56 million years ago), Eocene (56–34 million years ago) and Miocene (23–5.3 million years ago).
The models classified existing pollen accurately over 80% of the time and also showed high consensus on the identification of fossil pollen specimens. These results support previous hypotheses suggesting that Amherstieae originated in Africa and later dispersed to South America, revealing an evolutionary history of nearly 65 million years.
3D reconstruction of a pollen grain of Anthonotha showing external morphology. Credit: Ingrid Romero.
“We do not know the biological affinity of the majority of types of deep-time fossil pollen,” said STRI paleontologist Carlos Jaramillo, co-author of the study. “This study shows that with the right tools, we are able to taxonomically classify fossil pollen beyond what has been previously possible.”
However, over a third of the fossil specimens did not present biological affinity with any existing genera, suggesting that part of this ancient diversity may have went extinct at some point during the evolutionary process.
“These new tools reveal the vast amount of taxonomic information that pollen can offer and that has been hidden from researchers until now,” said Ingrid Romero, doctoral student at the University of Illinois-Urbana Champaign and lead author of the study.
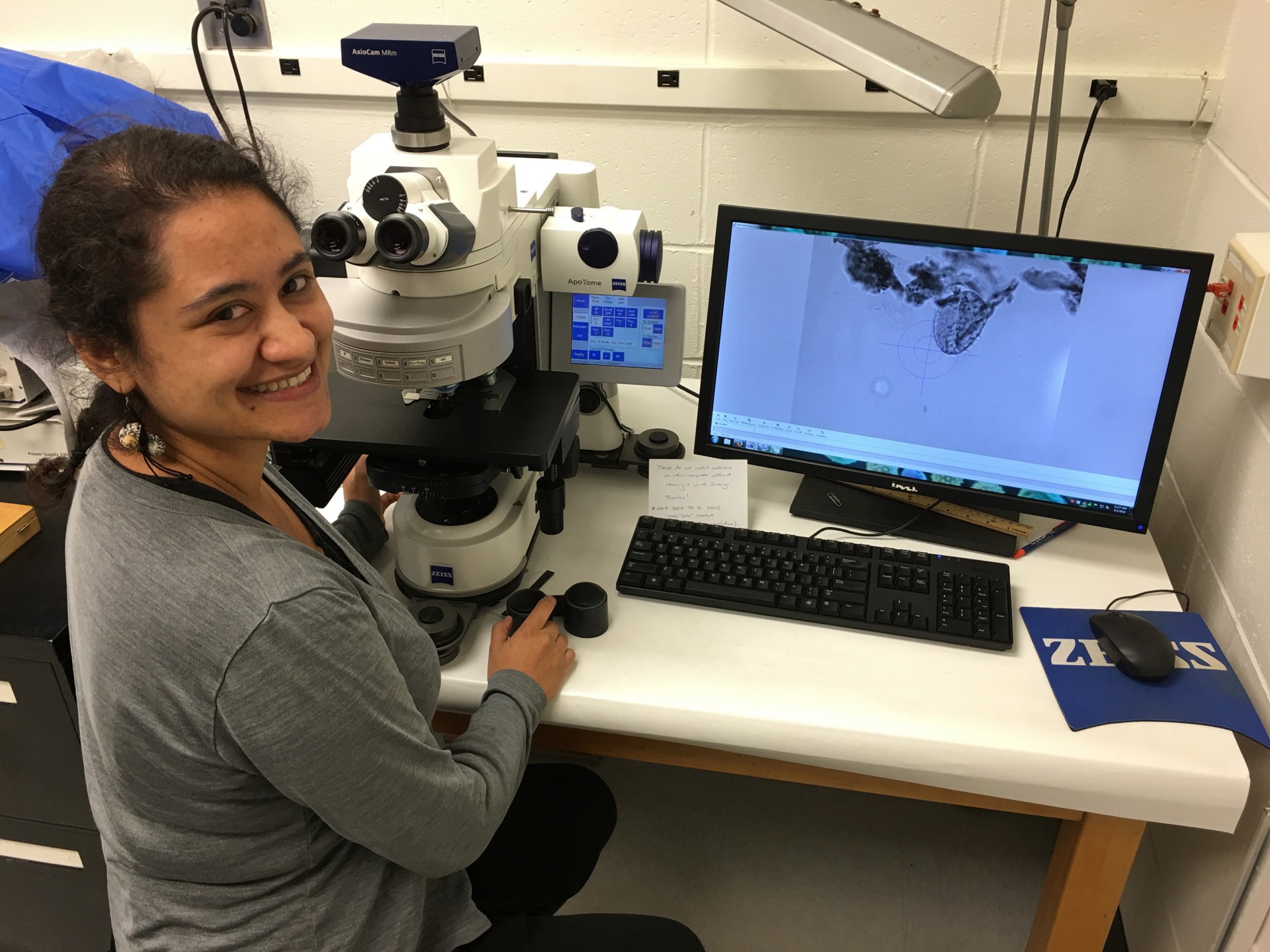
Researcher Ingrid Romero looking for fossil pollen in a light microscope. Credit: Doug Peterson.
This new approach improves the taxonomic resolution of fossil pollen identifications and greatly enhances the use of pollen data in ecological and evolutionary research. It also narrows down the range of options for experts in fossil pollen identification, allowing them to save time and invest their energy on the most challenging specimens.
“Fossil pollen analysis is a very visual science and computer vision algorithms augment expert identifications and can assist palynologists with challenging visual classification problems,” said Surangi Punyasena, associate professor at the University of Illinois-Urbana Champaign and senior author of the study. “The machine-learning models that our colleagues Shu Kong and Charless Fowlkes developed are truly remarkable. Our hope is that other researchers apply these techniques and that as a community we expand the ecological and evolutionary questions that are addressed by the fossil pollen record.”
Romero, I.C., Kong, S., Fowlkes, C.C., Jaramillo, C., Urban, M.A., Oboh-Ikuenobe, F., D’Apolito, C., Punyasena, S.W. (2020). Improving the taxonomy of fossil pollen using convolutional neural networks and superresolution microscopy. Proceedings of the National Academy of Sciences of the United States of America. https://doi.org/10.1073/pnas.2007324117