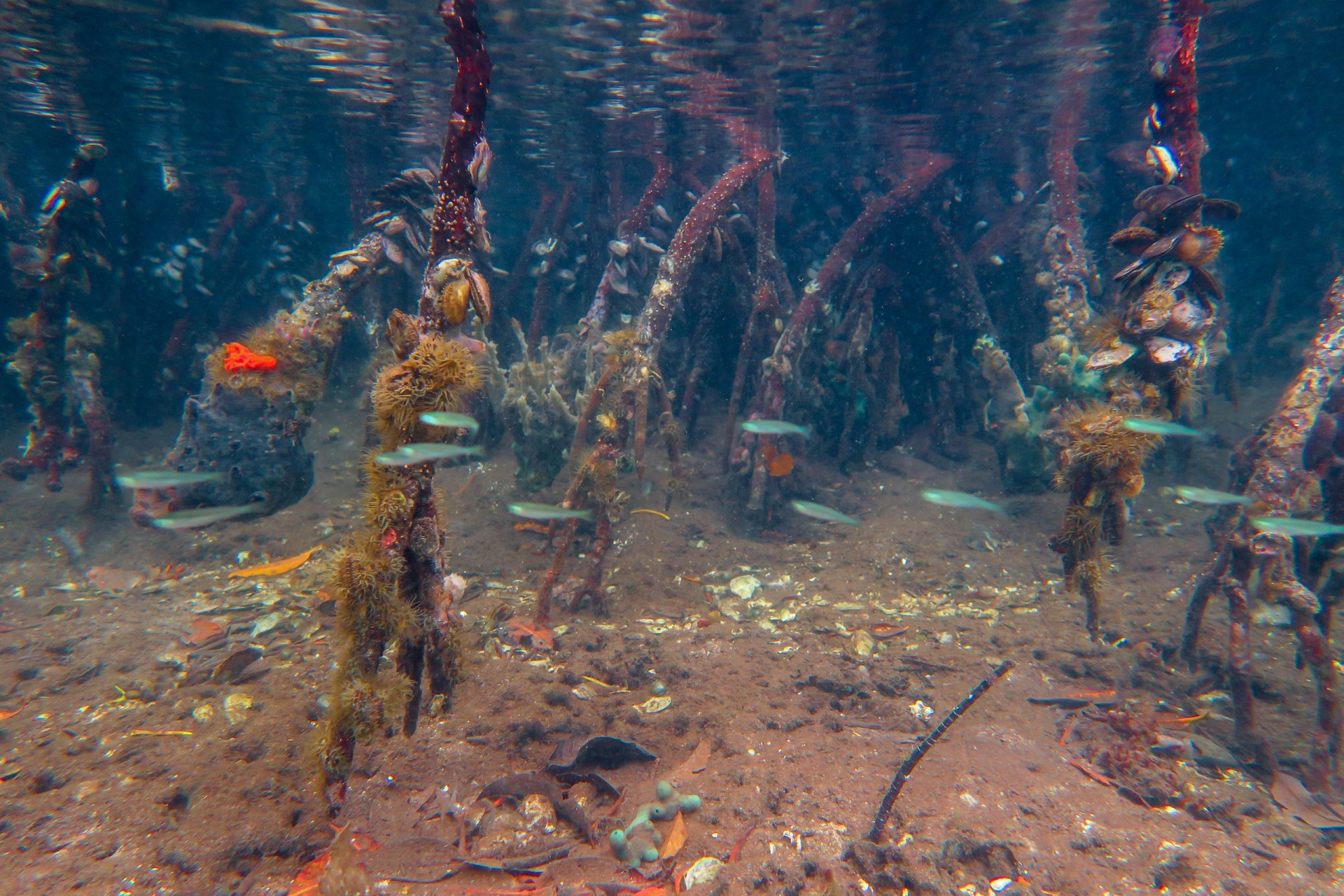How katydid songs expand
researchers’ knowledge of
tropical insects
All the fish
in the sea?
Smithsonian sleuths distinguish tropical
marine habitats in Panama using eDNA
Bocas del Toro
One of the big questions about using DNA in seawater to make species lists is whether it comes from a specific site or has floated in from elsewhere. In this study researchers could distinguish different marine habitats using only DNA.
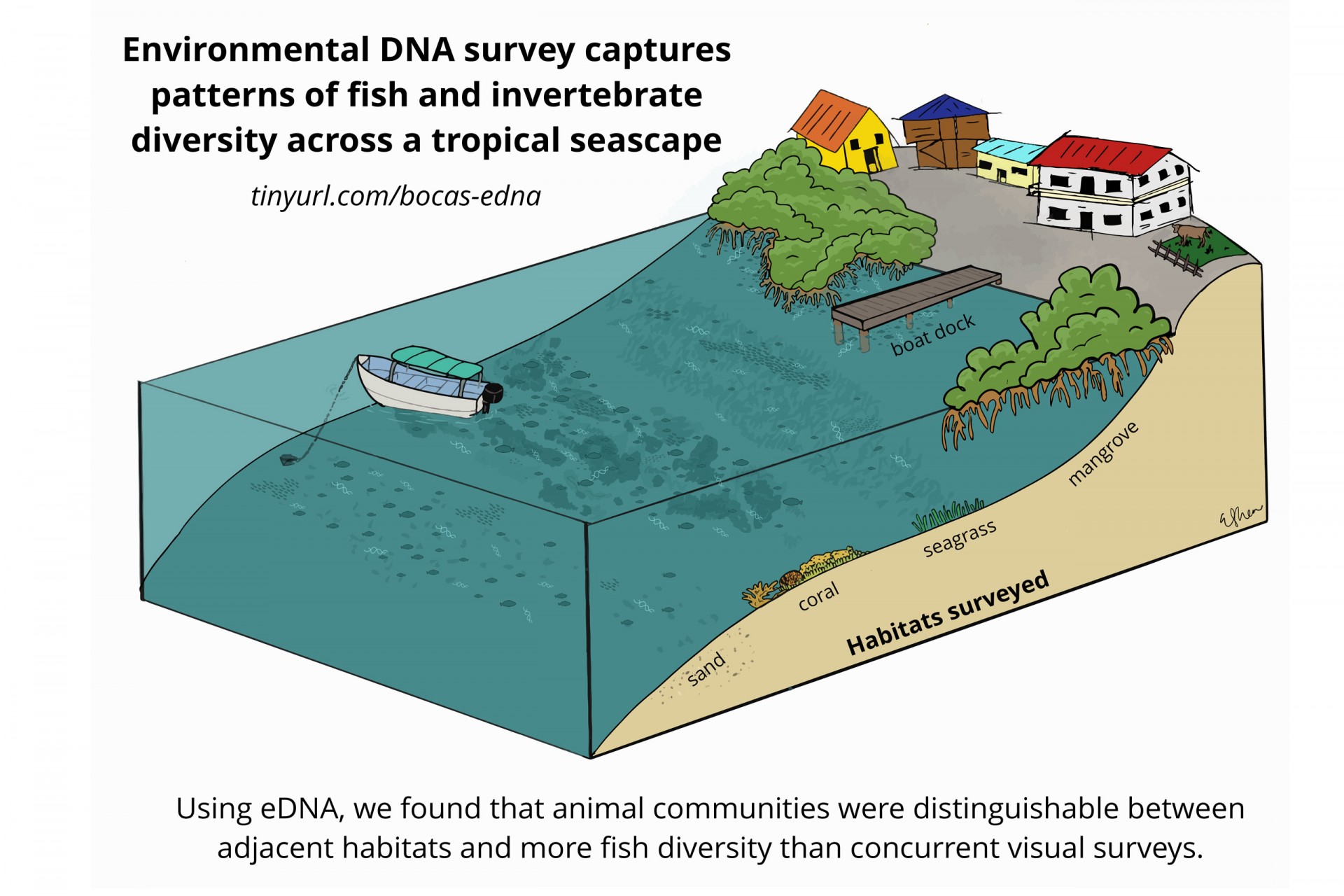
Researchers from six universities and museums met at the Smithsonian Tropical Research Institute (STRI) and discovered DNA from nearly 9000 different types of animals in the waters in and around Almirante Bay in Panama's Bocas del Toro Province. They bet on a powerful technique not only to identify endangered and invasive species, but also to monitor environmental changes in the Bay. The team showed that they could tell the difference between five different habitats based on minuscule bits of DNA floating in sea water. Not only did they detect many of the fish in the sea, they also detected thousands of marine invertebrates, microbes and even a mouse!
"Usually marine surveys focus on one group: fish, or invertebrates, or microbes," said Matthieu Leray, post-doctoral fellow at STRI, who leads a larger project to compare marine organisms in Panama's Caribbean and Pacific Oceans. "In this survey we tried to look at all animal groups simultaneously. One reason we could do that is because the marine fauna of Bocas del Toro is relatively well known."
From Left to Right: Elaine Shen, Will Wied, and Dr. Janina Seemann in the field collecting samples. While Elaine took water samples, Will and Dr. Seemann surveyed the fish communities at each site.
They compared five different habitats (mangroves, seagrass beds, sandy bottoms, coral reefs and boat docks) in the traditional way—a diver swims along and records all of the fish s/he sees—and also by collecting environmental DNA (eDNA) in water samples from each habitat.
Elaine Shen, then an undergraduate intern from Rice University, wrote the resulting paper in Scientific Reports based on her summer internship research project.
"I hope this inspires other undergraduates to apply for research funding and paid internships that seem out of reach, but they are actually attainable. It made me believe that, if you're willing to put in the work, anything is possible." Elaine, now a PhD student at the University of Rhode Island, is patiently waiting for the lockdown to end so that she can set off to Indonesia to work on her doctoral research using the same eDNA techniques in an area where much less is known.
Summary figure of the paper's main results (Elaine Shen).
DNA in seawater comes from whatever animals leave behind: poop, skin and blood cells, scales and mucus.
Geneticists determine and compare the sequence of nucleotides in each strand of DNA—imagine a necklace of different colored pearls. Each, unique sequence (pattern of colors) is called an operational taxonomic unit (OTU). By comparing these patterns to the DNA sequences from species collected before, they figure out which species were in each water sample. Because the Smithsonian has a marine station in Bocas, they took advantage of a huge database of 6,500 species that scientists have recorded during the last two decades as well as several other tools like a virtual guide to Shorefish of the Greater Caribbean, available as an iPhone app.
In total, they found 23,123 different OTU's. 8,781 of these were multicellular animals. And of these, they were able to identify about 800 species. Fish were less than one percent of the total, based on the eDNA results. Of all the OTU's that they could match with known species, only 40 percent were previously reported. That means that hundreds of new species are waiting to be discovered.
Though many shallow-water habitats of Bocas del Toro were close to each other in distance, they harbored different sets of taxa that drove biodiversity patterns within them.
Even though the divers who did the traditional, visual survey observed an estimated 7 million fish in total, the total number of species on their lists was only 97. The eDNA technique, which is less labor-intensive found 43 additional species.
By comparing the DNA from seawater, they could distinguish between mangroves and seagrass sites, between reefs and sandy sites and between sites that were in the bay versus sites exposed to open ocean conditions. Mangroves were the most diverse, followed by seagrass beds, open water (dock samples), sandy bottoms and coral reefs. Although the reefs may be home to the most conspicuous animals...brightly colored fish and sea anemones, these other habitats are home to inconspicuous animals that have yet to be identified.
"Our finding that eDNA collected from different habitats was different is important," Leray said. "There is no magic tool for studying marine biodiversity. People have been concerned that DNA in sea water may move around and may not represent a particular habitat. DNA lasts anywhere from 8 hours to a day, so it may move around to some extent, but we showed that it was specific to a given habitat."
"That was actually the coolest thing about our project," said Bryan Nguyen, fellow at the National Museum of Natural History in Washington. "Because we could show differences between habitats, we found out something new about eDNA that will be useful in the future as we monitor diverse seascapes. It also taught us the value of using both eDNA and visual methods. Each technique gives you different parts of the picture. With both, you get a much more complete view."
Nguyen BN, Shen EW, Seemann J, Correa AMS, ODonnell JL, Altieri AH, Knowlton N, Crandall, KA, Egan SP, McMillan WO, Leray M. 2020. Environmental DNA survey captures patterns of fish and invertebrate diversity across a tropical seascape. Scientific Reports. https://doi.org/10.1038/s41598-020-63565-9